CUTANA CUT&Tag Assays
CUTANA™ CUT&Tag: Ultra-low input chromatin mapping
Cleavage Under Targets and Tagmentation (CUT&Tag) is a novel immunotethering technology, based on Cleavage Under Targets and Release Using Nuclease (CUT&RUN) methods.
In CUT&Tag, a fusion of protein A, protein G, and Tn5 transposase is used to catalyze simultaneous cleavage and sequencing adapter ligation at antibody-bound chromatin. The use of Tn5 in CUT&Tag is a key advance, as this step eliminates the need for chromatin fragmentation as well as expensive library preparation.
CUT&Tag assays display significantly improved signal-to-noise (S/N) vs. ChIP-seq assays, particularly for mapping histone PTMs in ultra-low sample inputs.
CUTANA CUT&Tag Kits
The CUTANA CUT&Tag Kits offer a comprehensive solution for ultra-sensitive mapping of histone post-translational modifications (PTMs). These kits use an exclusive Direct-to-PCR strategy to go from cells to PCR-amplified sequencing libraries in one tube, bypassing traditional library prep and minimizing sample loss. The recommended input for CUT&Tag is 100,000 native nuclei per reaction. Comparable data can be generated down to 10,000 nuclei, and the protocol is also validated for whole cells, cryopreserved samples and lightly cross-linked nuclei or cells.
- Cells to sequencing in 2 days
- Exclusive single-tube protocol, no library prep
- Designed for multi-channel pipetting and 8-strip tubes
- Reliable data down to 10,000 cells
- Only 5-8 million sequencing reads
- All necessary reagents included
How it works
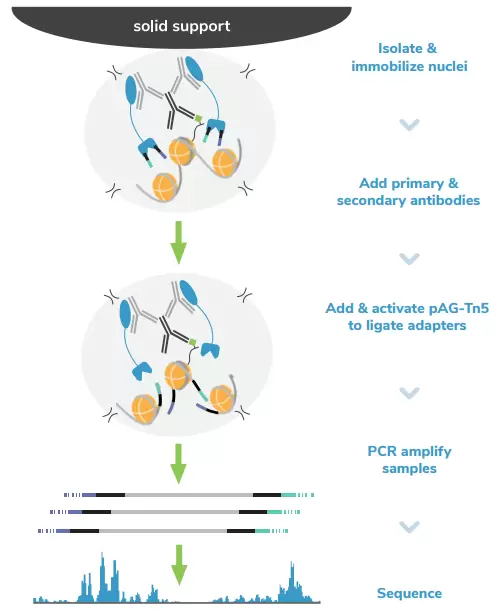
CUTANA CUT&Tag Workflow. In CUTANA CUT&Tag, nuclei are immobilized to a solid support and antibodies bind their chromatin targets in situ. A fusion of proteins A and G with prokaryotic transposase 5 (pAG-Tn5) is used to selectively cleave and tagment antibody-bound chromatin with sequencing adapters. Tagmented fragments are directly PCR-amplified using EpiCypher’s exclusive single-tube (“Direct-to-PCR”) approach, yielding sequence-ready DNA.
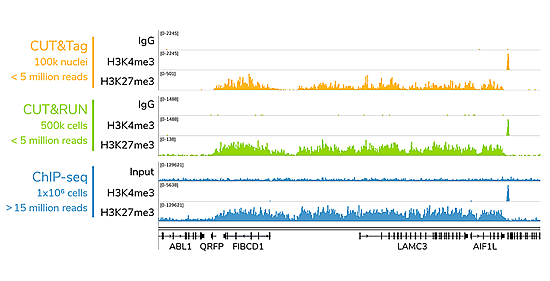
Quality data with lower input and sequencing depth requirements
CUTANA CUT&Tag generates similar results at a fraction of the cell input and sequencing depth used in ChIP-seq. A representative 300 kb region at the LAMC3 gene is shown for CUT&Tag (orange), CUT&RUN (green) and ChIP-seq (blue) data generated using H3K4me3 and H3K27me3 antibodies (EpiCypher 13-0041 and 13-0030, respectively). Rabbit IgG Negative Control Antibody (EpiCypher 13-0042) and ChIP Input control are shown for comparison (control tracks are scaled to the track with the highest signal in each approach).
Components for CUT&Tag Set Ups
CUT&Tag Kits & Core Reagents
CUT&Tag compatible Antibodies
CUTANA™ Quick Cleanup DNA Purification Kit
References
- Kaya-Okur et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Comm. 10, 1930 (2019). (PMID: 31036827)
- Kaya-Okur et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264-3283 (2020). (PMID: 32913232)
- Henikoff et al. Efficient transcription-coupled chromatin accessibility mapping in situ. by nucleosome-tethered tagmentation. Elife 16;9:e63274.(2020). (PMID: 33191916)