Single-cell RNA-seq
Single Cell RNA Sequencing
Microfluidics-free, templated emulsification-based scRNAseq technology - flexible, scalable, and affordable
Single-cell RNA sequencing (scRNA-Seq) has enabled unprecedented insight into the biology and pathology of individual cells across a broad range of discovery and disease applications. However, specialized capital equipment, high reagent costs, lack of accessibility, and scalability are key factors limiting the wide-scale adoption and use of single-cell technologies.
Therefore, our partner Fluent BioSciences has developed the PIPseq 3’ Single Cell RNA product line based on a novel technology, published in Nature Biotechnology, which uses a simple vortexer and standard lab equipment to enable researchers to conduct single-cell RNA sequencing studies affordably and at a wide range of scales.
- Easy to implement, no complex instrumentation or consumables
- Highest cell capture rate in the market (up to 80%)
- Flexibility to process any number of reactions per kit as needed
- Cost-effectively scale from pilot and low cell diversity projects to complex tissue analysis all with the same technology
- Conveniently and quickly process cells and capture RNA at point of collection
- < 10 min of hands-on time until first stable stopping point
- User-friendly PIPseeker software for downstream analysis
PIPseq Technology
Fluents novel approach relies on Particle-Templated Instant Partitions (PIPs) to simultaneously segregate complex cell mixtures into partitions with barcoded template particles that can be easily processed for single cell applications such as single cell RNA sequencing (scRNA-Seq). This approach (PIPseq™) eliminates the need for complex, expensive instrumentation and microfluidic consumables.
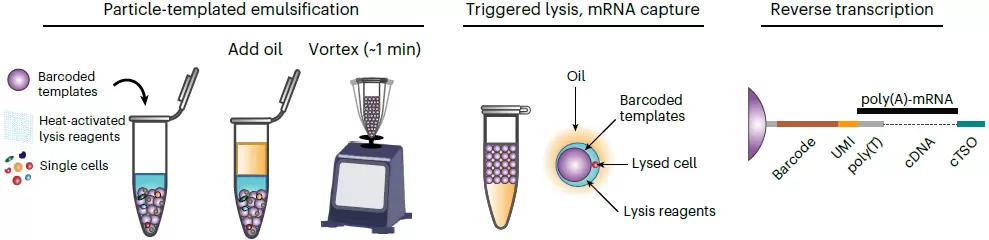
Workflow
Sample Preparation
- During sample preparation, cell suspension of interest is mixed with the core template particles and segregated into Particle-Templated Instant Partitions (PIPs) by simple vortexing
- The cells in PIPs are then lysed on a thermal device and the mRNA is captured by barcoded oligonucleotides incorporated with the template particles
Library Preparation
- The captured mRNA undergoes reverse transcription and cDNA libraries are then generated followed by adapter ligation
- Sample indices (compatible with Illumina sequencing) are then added, followed by a final cleanup
Next generation sequencing (NGS)
- Libraries can then be processed on the appropriate Illumina NGS instrument. The sequencing reagent kit (and chip) is chosen based on the number of samples multiplexed, cell count and desired read depth
Data Analysis
- The Fluent PIPseekerTM Software enables primary analysis of the PIPseq sequencing libraries
- Upload your FASTQ files into the software and obtain summary metrics, diagnostic plots, clustering and differential gene expression tables
- Also generate standard feature-barcode count matrices, which are compatible with widely-used open-source tertiary analysis packages