DriverMap Adaptive Immune Receptor Profiling
DriverMap Adaptive Immune Receptor (AIR) TCR-BCR Profiling
Profile the full-length receptor region or only the CDR3 repertoire of all 7 TCR and BCR chains in a single-tube, single-day assay
DriverMap AIR is the only assay technology on the market that profiles the full-length receptor region (exclusively RNA) or only the CDR3 repertoire of all T-cell receptor (TCR) - TRA, TRB, TRG and TRD - and B-cell receptor (BCR) - IGH, IGK and IKL - chains in an single-tube, single-day assay using multiplex PCR-NGS technology without the need of any additional specialized equipment. Cellecta offers AIR assays to profile human or mouse RNA, DNA, or both. The simultaneous profiling of DNA and RNA enables the identification of antigen-activated clonotypes to provide even greater insight into the immune response.
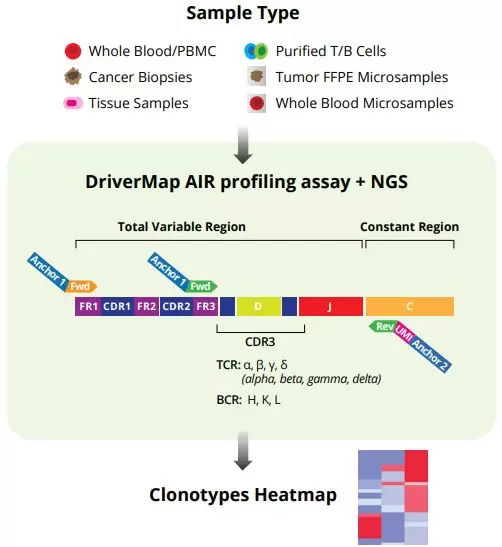
DriverMap Multiplex PCR technology uses gene-specific primers which significantly reduce the level of non-specific binding and primer-dimer amplification products and are designed to target only expressed TCR/BCR mRNA isoforms. Unique Molecular Identifiers (UMIs) facilitate accurate quantitation of the copy number of cDNA molecules in amplification steps, as well as detection of low abundance clonotypes and correction of amplification biases and sequencing errors. Furthermore, the dual-index amplicon labeling strategy minimizes index hopping during NGS allowing for comprehensive readouts.
While the DriverMap AIR-RNA assay quantifies T-cell and B-cell receptor transcripts (see Fig.2.) the DriverMap AIR-DNA assay amplifies receptor genes directly from genomic DNA. Combining data obtained from both the AIR-DNA and AIR-RNA assays enables assessment of both the transcriptional activation and number of cells with a particular clonotype. The ability to differentiate these two effects provides a quantitative basis to assess antigen-activated clonotypes.
How It Works
- Start from RNA or DNA from a variety of immune sample types such as whole blood, PBMCs, cancer biopsies, tissue samples, FFPE or dried blood microsamples
- Detect and accurate quantify AIR clonotypes with the multiplex PCR-based assay making use of Unique Molecular Identifiers (UMIs)
- Obtain more sensitive and reproducible profiling data
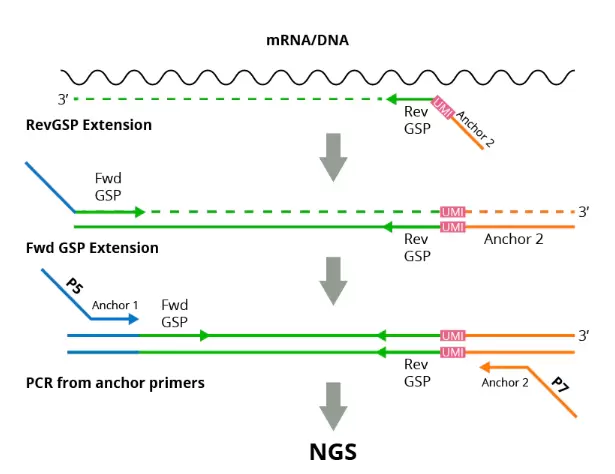
Fig.1. Workflow of RNA/DNA assay.
DriverMap AIR differs from other Adaptive Immune Receptor Repertoire (AIRR) Assays
- DriverMap™ Multiplex PCR technology uses gene-specific primers which significantly reduce the level of non-specific binding and primer-dimer amplification products, and are designed to target only TCR/BCR isoforms.
- Unique Molecular Identifiers (UMIs) facilitate accurate quantitation of the copy number of cDNA or DNA molecules in amplification steps, as well as detection of low abundance clonotypes and correction of amplification biases and sequencing errors.
- Dual-index amplicon labeling strategy minimizes index hopping during NGS allowing for comprehensive readouts.
Full profiles of the antigen-recognition CDR3 region enable assessment of CDR3 length distribution, V(D)J segment usage, isotype composition for BCRs, somatic mutations, and similar characteristics with immune receptor profiling software such as MiXCR (MiLabs).
Applications of BCR Sequencing
- Identify broadly neutralizing antibodies (BNAbs) and map Ig-seq datasets to known antibody structures for antibody and vaccine development
- Track B-cell migration and development patterns
- Find clinical markers of autoimmune diseases such as multiple sclerosis, rheumatoid arthritis and cancers (e.g. B-cell lymphoma)
- Contrast naïve and antigenically challenged datasets for understanding antibody maturation
Applications of TCR Sequencing
- Track T-cell clonality and diversity for mechanism of action of immune checkpoint inhibitors for immunotherapies
- Assess TCR overlap between repertoires to define spatial and temporal heterogeneity of the anti-tumoral immune response
- Analyze TCR sequence and structure to annotate antigenic specificity for developing personalized cellular immunotherapies
Supporting Data
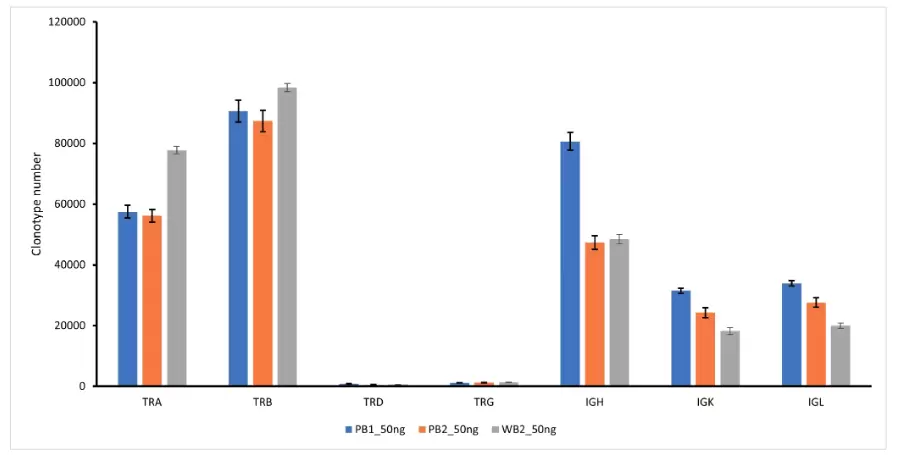
Fig.2. Number of clonotypes for 7 TCR/BCR chains identified in 50ng of normal PBMC or whole blood RNA (10x10^6 reads per sample, triplicates).
Product Flyers and Other Resources
- Flyer: Comprehensive Adaptive Immune Receptor Profiling for All Immune Sample Types
- Video: Improved Adaptive Immune Receptor Repertoire Profiling for Biomarker Discovery (23:08)
DriverMap Adaptive Immune Receptor (AIR) TCR-BCR Profiling Service
Do you have immune samples you would like to profile? Provide us with some information on your samples and your study goals, and we will get back to you to discuss your project.