DriverMap Targeted Expression Profiling
DriverMap Targeted Expression Profiling (EXP)
Genome-wide, targeted expression analysis that combines the sensitivity of multiplex PCR with the dynamic range and throughput of NGS
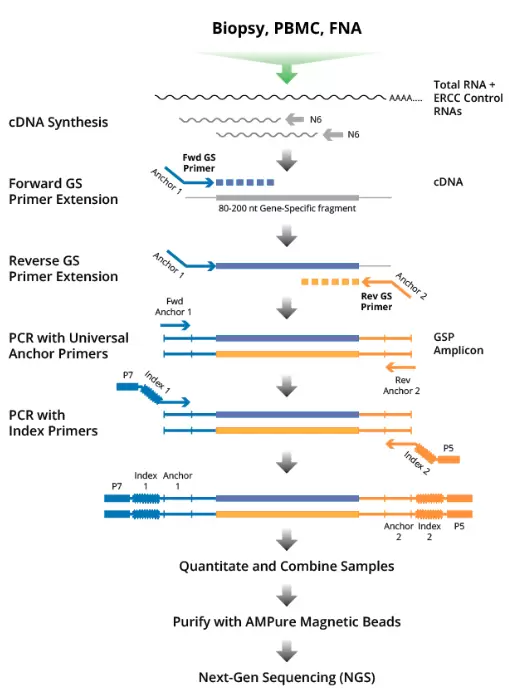
The DriverMap Targeted Expression Profiling Platform enables targeted multiplex PCR amplification and next-generation profiling to measure the expression levels of up to 19,000 human protein-coding genes in a single assay. By combining highly multiplexed RT-PCR amplification with the depth and precision of next-generation sequencing (NGS) quantitation, the DriverMap assay provides convenient, comprehensive, highly sensitive, and quantitative measurement of gene expression from as little as 10 pg of total RNA.
Available off-the-shelf assay formats include the DriverMap Human Genome-Wide Expression Profiling Kit and a number of panels of expertly curated gene targets, see table below.
A DriverMap Targeted RNA Sequencing Service is also offered.
- Single-tube, one-day protocol starting with as little as 10 pg total RNA to NGS library for sequencing with next-generation sequencing (NGS) readout
- Higher sensitivity for low expressed genes than RNA-Seq with 10-fold less sequencing depth required
- Simplified analysis against reference sequences - genomic alignment not needed
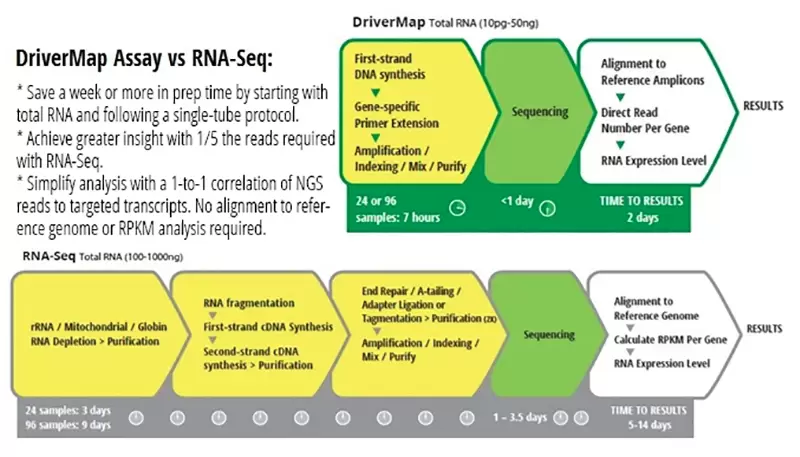
How It Works
Rather than reverse-transcribing the whole transcriptome, the DriverMap assay combines multiplex RT-PCR amplification to amplify defined and conserved 80- to 200- base regions of the targeted genes, and then uses next-generation sequencing (NGS) to quantitatively assess abundance levels of each of these transcript amplicons.
There are significant advantages to targeting, amplifying, and sequencing carefully selected discrete loci of interest in the expressed transcriptome. Gene-specific primers that target each gene of interest obviate the need for RNA preparation to remove unwanted sequences such as ribosomal RNA, beta-globins, or other non-coding RNA. Only transcript sequences matching the targets of interest are amplified. Therefore, only a small amount of total RNA is needed for this highly sensitive assay. As little as 10 pg of total RNA from single-cell lysate is enough to detect most target transcripts.
Also, the NGS read depth required to reliably read several thousand targeted amplicons is much less than the depth required for the whole transcriptome. As a result, the targeted approach detects lower-abundance expressed transcripts more consistently and reliably than other RNA-seq approaches and much less sequencing depth is required.
Analysis of the sequencing results is much simpler with the DriverMap targeted approach since each amplicon corresponds to a known reference sequence. There is no need to estimate gene copy number from assembled cDNA fragments. With targeted RT-PCR the read levels of each amplicon directly correlate to the expression levels of the target transcript. The analysis can be done entirely on an Excel spreadsheet using housekeeping genes as references in a manner similar to standard qRT-PCR. Software is included with the DriverMap kit to enable this amplicon alignment and to generate a spreadsheet of read counts for targets directly from the Illumina FASTQ file.
DriverMap Assay vs RNA-seq